
During the process of meiosis, which is necessary for sexual reproduction, DNA breaks occur, and repairs take place. PARG-1, a key regulator, plays a role in maintaining genome integrity and offers insights relevant to human health. Thanks to a Standard Grant provided by The Czech Science Foundation to Nicola Silva at Masaryk University, he was able to explore the complexities of DNA repair dynamics in the germ line.
Human cells consist of 46 chromosomes, half of which are received from our mother and the other half from our father. During development, all sexually reproducing organisms undergo a process called meiosis, in which chromosome number is reduced by half into the newly formed germ cells, oocytes and sperm. Upon fertilization, the genetic information carried by oocyte and sperm fuses into the zygote, thus reconstituting the original number of chromosomes.
Several mechanisms exist in meiosis to ensure that each gamete, sperm and oocyte, receives the right number of chromosomes. If the process is inaccurate and leads to the production of dysfunctional gametes, it can result in the transmission of inheritable mutations to the offspring.
One key aspect of meiosis is the exchange of physical DNA sequences within each pair of maternal and parental homologous chromosome in a process called Homologous Recombination. DNA lesions called double-strand breaks (DSBs) are created, maternal and parental chromosomes physically connect and exchange DNA sequences to achieve repair of these breaks, resulting in the formation of a crossover.
Crossovers are essential for faithful segregation of chromosomes into the daughter cells, and at the same time they lead to reshuffling of genetic traits, promoting genetic diversity. However, the number of breaks produced during meiosis in the germ cells greatly surpasses the final number of crossovers, indicating that repair systems that use homologous sequences to restore genome integrity can also produce non-crossovers events.
Tight regulation of the number of DNA double-strand breaks
DNA interruptions carry an intrinsic threat to genome integrity, therefore their number, placement across the genome, as well as the activation of the DNA repair system(s) in charge to resolve them, must be tightly regulated. Several genes involved in the regulation of DNA repair have been identified over the years and mutations in virtually any of these contribute to the development of cancer-prone syndromes in humans, such as breast/ovarian cancer susceptibility genes BRCA1/BRCA2, Fanconi Anaemia, and many others.
One way of controlling DNA repair is through the attachment of chemical groups to a protein after synthesis or the removal of signal peptides after cellular localization in a process called post-translational modification. One of the major changes that occur in response to DNA damage is Poly(ADP)ribosylation (PARylation), a process in which ADP-ribose units are added to substrates to regulate their activity. Although extensively studied in ex vivo models, examining PARylation in living organisms has been challenging in mammals due to the embryonic lethality associated with the complete loss of function mutations in its “writers,” PARP1/2. These enzymes are responsible for synthesizing ADP-ribose chains in response to genotoxic stress. Additionally, the “eraser” enzyme PARG, which counteracts PARP1/2 activity by breaking down ADP-ribose chains, further complicates in vivo analysis.
The non-parasitic nematode Caenorhabditis elegans, a renowned model system for genome stability studies, confers a tremendous advantage compared to other models since loss-of-function mutations in both PARP1/2 or PARG are tolerated, allowing study of their function within the germ line.
Thanks to a Standard Grant provided by The Czech Science Foundation to Nicola Silva at the Department of Biology of the Faculty of Medicine, Masaryk University, an in-depth and unprecedented in vivo analysis of the roles exerted by these proteins could be carried out, leading to the identification of crucial functions performed particularly by PARG-1 (C. elegans homolog of mammalian PARG) during meiosis.
By exploiting genome editing techniques (CRISPR), we were able to assess PARG-1 localization in developing oocytes, a task that was never carried out before. Our analysis revealed that PARG-1 is an integral component of an important meiotic scaffold that keeps the chromosomes tightly aligned during meiosis, called the Synaptonemal Complex (SC) (Figure 1).
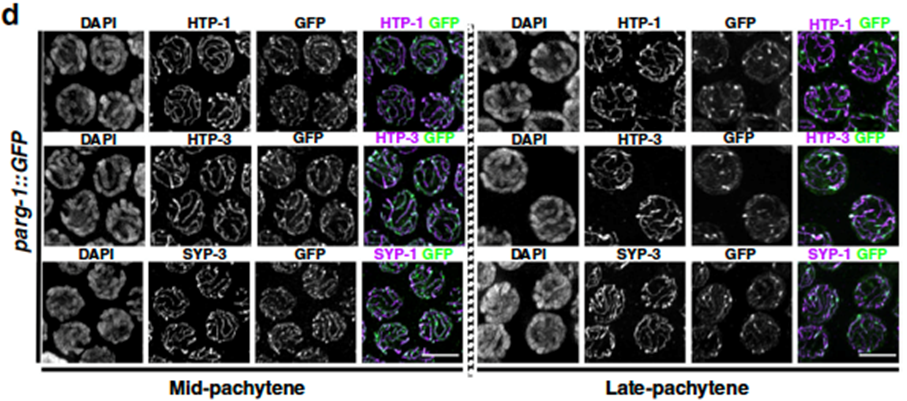
Figure 1. Oocytes at the indicated stage stained for different subunits of the SC and PARG-1 (GFP). From Janisiw et al.; Nature Communications, 2020.
Moreover, we were able to find that PARG-1 establishes protein complexes also with proteins involved in the induction and processing of meiotic DSBs across species. By exploiting the powerful genetics provided by C. elegans, we discovered that strikingly PARG-1 is important to regulate the number of DNA breaks during germ cells development, and to promote their precise repair by homologous recombination (Figure 2).
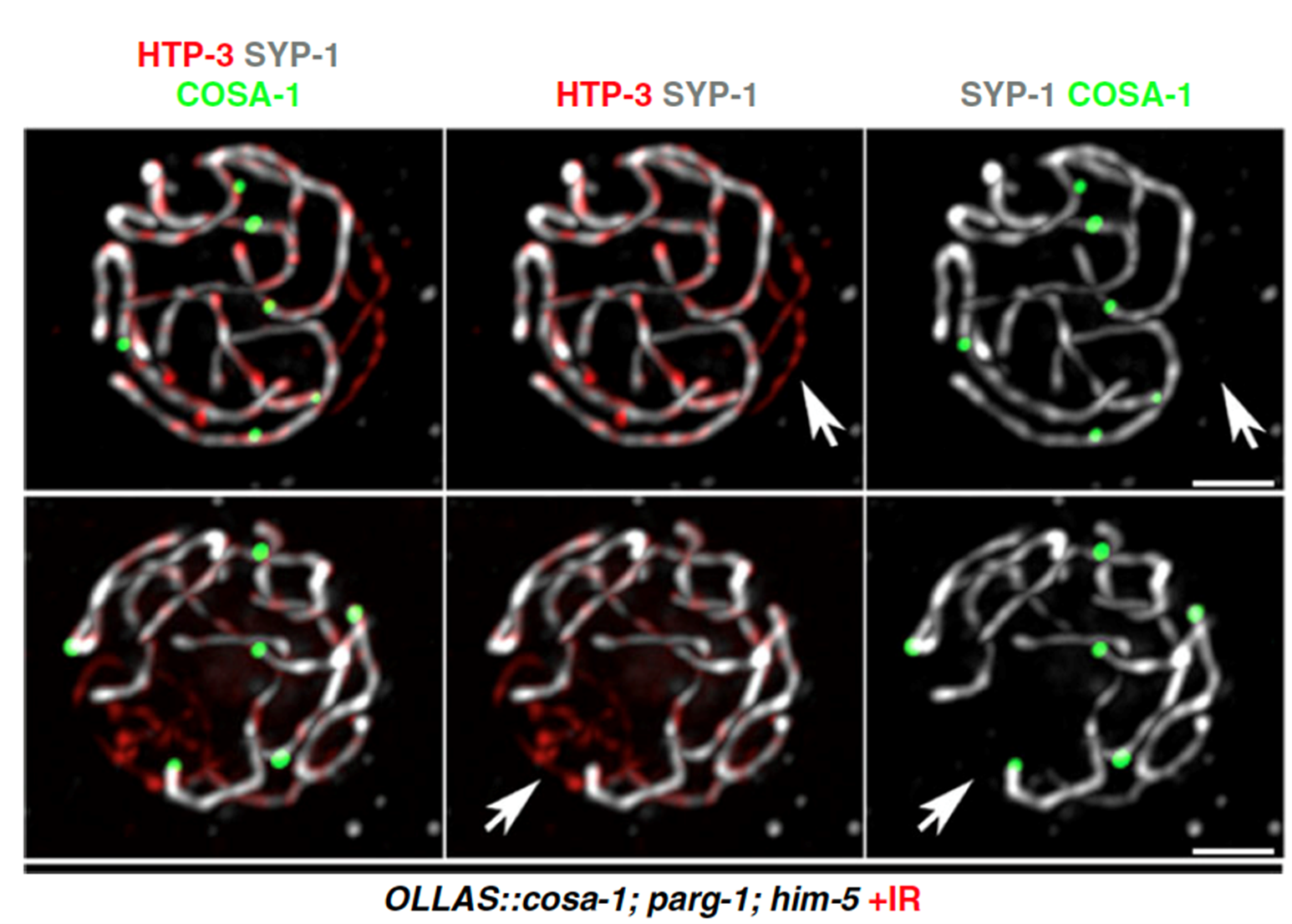
Figure 2. Oocytes stained for different SC subunits and the crossover sites in mutants with reduced DSBs, after irradiation. From Janisiw et al.; Nature Communications, 2020.
Lastly, we discovered that the ability of PARG-1 to localize along the chromosomes holds crucial roles during DSB induction and homologous recombination, indicating that not only PARG-1 carries out enzymatic functions but is also important from a structural perspective.
We were also interested in deciphering the protein network established by PARG-1 and its interactors within the germ cells. This analysis led to the identification of a physical and functional interaction with the BRC-1-BRD-1 complex, which in humans is homologue to BRCA1-BARD1. These proteins perform major roles in regulating genome stability in mitotic cells, however their functions during meiosis are more elusive. Our work revealed that contemporaneous removal of PARG-1 and BRC-1 caused elevated infertility due to extensive genome instability. In particular, we observed that many DNA lesions physiologically generated during meiosis were not processed correctly, leading to the formation of chromosome fusions and reduced levels of recombination.
By removing different DNA repair pathways together with BRC-1/PARG-1 we observed detrimental effects on genome integrity caused by lack of the polymerase POLQ-1 (Figure 3), known to mediate an important DNA repair pathway called “alternative non-homologous end joining”.
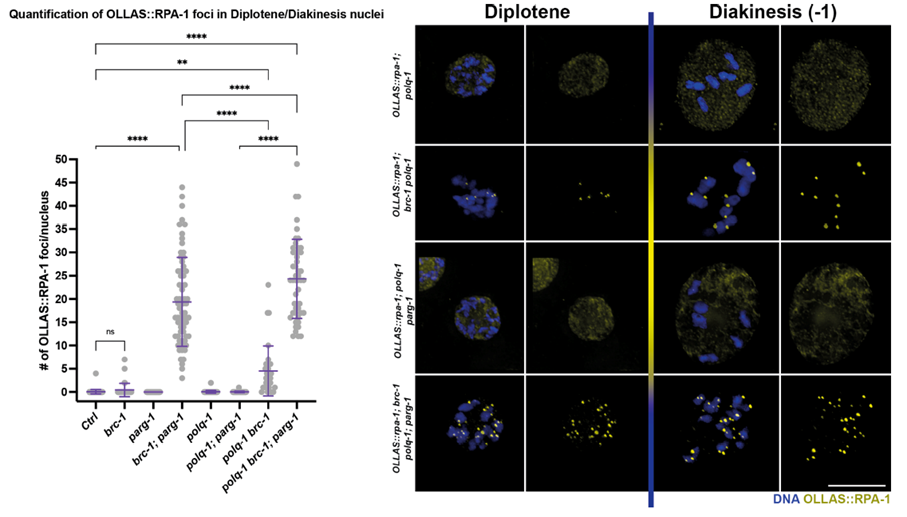
Figure 3. Oocytes of the indicated genotype and stage stained for a marker of single-stranded DNA, highlighting presence of unrepaired damage in absence of BRC-1-PARG-1. From Trivedi et al.; Nucleic Acids Research, 2022.
Our work was the first in vivo evidence showing that under compromised BRC-1/BRCA1 function, PARG-1/PARG activity is essential to maintain genome integrity in gametes. Furthermore, our finding on the synthetic lethality triggered by simultaneous POLQ-BRCA1 abrogation of function also carries tremendous ramifications for cancer therapy and nicely aligns with recent findings in humans that show promising results in targeting tumorous cells with POLQ inhibitors as a novel strategy to improve therapeutic outcomes in BRCA1-mutated tumours. Altogether, our work emphasizes the pivotal role that model systems carry for the analysis of conserved pathways in simpler organisms and further corroborates on the crucial contribution of basic science to human health.